Research Overview
The research in the Dearing lab focuses on understanding how small mammals overcome challenges related to diet and disease. Our work draws on approaches from many disciplines (e.g., physiology, ecology, pharmacology, genetics, biochemistry, ethology) and combines field and laboratory studies. Overviews of current projects are listed below – for additional details see individual pages of graduate students and postdoctoral fellows. Please see the publications page for a full listing of our previous work.
Understanding the genetic underpinnings that enable ingestion of poisonous diets
At every meal, animals that feed on plants (“herbivores”) face the possibility of being poisoned by naturally-occurring toxic chemicals in their food. Nevertheless, how herbivores process natural toxins in their regular meals remains poorly understood. For example, remarkably little is known about the specific genes that control an animal’s ability to process potentially toxic compounds in plants. To address these fundamental problems, this research focuses on a dramatic dietary change: woodrats normally eat juniper and cactus, but several populations in the American southwest have switched to a diet of creosote bush. Creosote bush produces natural toxins that radically differ from juniper and cactus, and therefore the specialized woodrat populations must detoxify their diets differently than the juniper and cactus-eaters. The goal of this project is to identify DNA-level changes that are associated with woodrats’ ability to feed on a toxic diet of creosote bush. This project will develop new genomic tools that will be useful to the broader scientific community. A better understanding of how animals process toxins can also impact decisions about pharmaceutical development for humans and other animals, and influence feeding options to improve production of free-ranging domestic herbivores. This work is a collaborative effort with Dr. Michael Shapiro (University of Utah) and Dr. Marjorie Matocq (University of Nevada, Reno). This work is funded by the NSF.
Investigating the role of gut microbes in facilitating the ingestion of dietary toxins
Recent studies have revealed that mammals are not individuals but rather are “superorganisms” that host diverse and interactive communities of microbes. Some of the most critical interactions facilitating mammalian life occur between mammals and the complex communities of microbes that reside within their guts. Plant-eating mammals harbor the most diverse microbial communities. Many of these microbes are important in the degradation of fiber; however, gut microbes may also play an essential role in detoxifying the natural toxins common in plants. This project investigates how host evolutionary history and dietary toxins shape the diversity of the gut microbiome of herbivorous mammals by focusing on a group of rodents (woodrats) that specialize on toxic plants. The objectives of this project are to 1) identify and compare the microbial communities across chambers of the woodrat gut to understand their function 2) investigate the influences of evolutionary history and dietary toxins in sculpting microbial diversity and function; 3) determine how diversity interacts with the liver of the host to facilitate detoxification. This project will provide some of the first insights into the microbial diversity of wild herbivores and the role that plant toxins play in shaping diversity. This collaborative effort includes Dr. Colin Dale and Dr. Robert Weiss (University of Utah), and Dr. Kevin Kohl (University of Pittsburgh) and was funded by a Dimensions of Biodiversity award from the NSF.
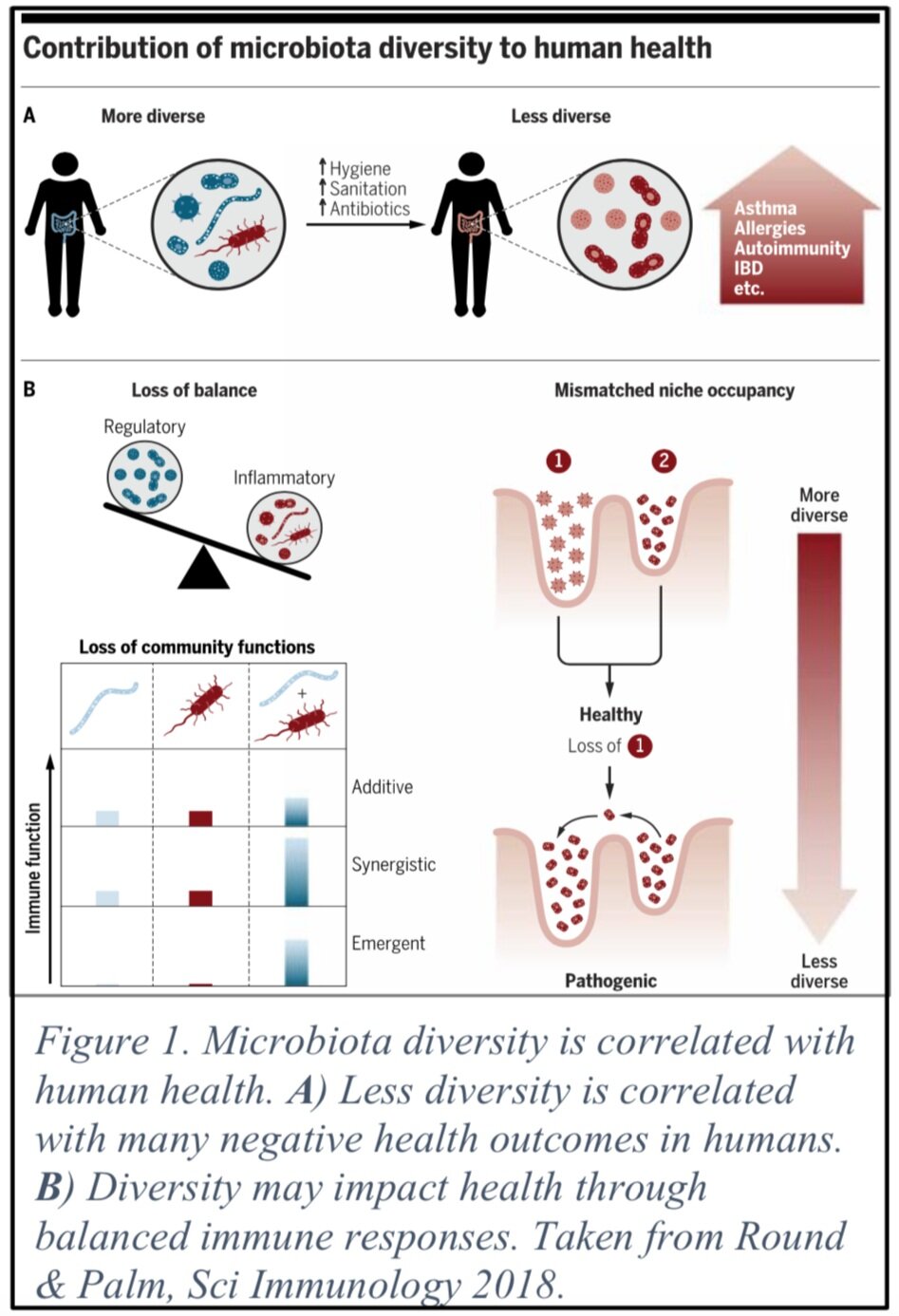
Rules of Resilience: Modeling impacts of host-microbe interactions during perturbations
The gut microbiota has emerged as an integral component of organism well-being, influencing the most basic of functions such as metabolism, to more specialized systems e.g., immunological function. While work has been done to understand the interactions of host, microbiota, and environment in one dimension, few have attempted to integrate all three of these aspects. This gap is largely due to a lack of computational tools that have the capability to integrate and build models from the large multi-omic datasets that arise from each component. Viewing the combined data as high dimensional tensors allows us to use techniques such as factor analysis and latent feature representations to find multi-way correlations across the three components. The increased dimensionality also necessitates the need for being able to perform such analyses in a scalable fashion using modern supercomputers, (e.g., NSF-funded Frontera). This work is a collaboration with Dr. June Round, Dr. Zac Stephens, Dr. Hari Sundar, and Dr. Aditya Bhaskara (University of Utah) and is funded through the 1U4U.